| Genome regulation & evolution |
 Research officer (CR1), Inserm
Research officer (CR1), InsermCette adresse email est protégée contre les robots des spammeurs, vous devez activer Javascript pour la voir. Tel: +33(0)540008404
Denis Dupuy initially trained in Biology at University of Pau and got his Master of Science in Molecular and Cell Biology at Université Bordeaux Segalen. He did his Ph.D. thesis in human genetics in the laboratory of Dr. Benoit Arveiler at the University of Bordeaux (1998-2001) working on positional cloning of schizophrenia susceptibility gene. He then joined the group of Dr Marc Vidal, at the Dana-Farber Cancer Institute (Harvard Medical School, Boston, Ma) for a Post-Doctoral training in systems biology. There he acquired the tools and methods needed to perform systematic analysis of spatiotemporal gene expression in vivo in C. elegans.
C. elegans, development, GFP, systems biology, bioinformatics, high throughput RNAi, in vivo, miRNA, splicing, post transcriptional regulation, evolution
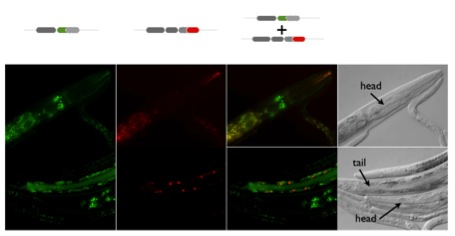
The major goal of our group is to generate an integrative model of tissue-specific post-transcriptional regulation processes in Caenorhabditis elegans. Many cis-acting elements and trans-acting factors involved in the regulation of these processes have been characterized. However, integrative models of the molecular mechanisms underlying the sophisticated cell- and stage-specific patterns of regulation are yet to be developed due to difficulties in following these events in vivo. Post-transcriptional regulation represents a critical aspect of genetic regulatory networks in eukaryotes. To dissect the genetic requirements for these mechanisms we will generate the first quantitative genome-scale dataset of post-transcriptional regulation in vivo during C. elegans development. Activity reportWe focus our effort on two major aspects of post-transcriptional regulation and in particular the analysis of alternative splicing
In summaryTo build dynamic models of cell differentiation it will be important to integrate comprehensive datasets of expression information and physical relationships between regulators and their targets within the system of interest. Tremendous efforts are underway to collect such datasets in C. elegans which make it the ideal model organism to reach this objective. Our goal is to complement these approaches with a systematic quantitative analysis of major spatiotemporal post-transcriptional regulation processes in vivo in C. elegans.
In 2012 D. Dupuy launched the first Bordeaux based team to participate in the International Genetically Engineered Machine (IGEM) competition. For the fourth participation a team of twenty eight students gathered every week for evening sessions of discussions and planning for a new synthetic biology project to be carried out over the summer. Fourteen undergraduate students represented the «Cured vine» project at the IGEM Giant Jamboree in Boston in September 2015 and were awarded a Gold medal in the Environment track.
|
2, Rue Robert Escarpit - 33607 PESSAC - France
Tel. : +33 (5) 40 00 30 38 - Fax. : +33 (5) 40 00 30 68